生物信息与R语言QQ群: 187923577
Tips: 看不清请刷新,换个颜色再看。
1. 比较干净的背景: +theme_bw(); 最干净的背景: +theme_classic()
2. 参数的解释: 生物慕课网
3. 本页面最底部有生信QQ群号,欢迎加入讨论,严禁广告。
#############################
library(ggplot2)
library(reshape2)
#示例数据:某基因在对照和肿瘤样本中的表达量。
d1=data.frame(
control=c(10,20,30,40,30,60,20,40,20,20,10,20,30,40,30,40,20,40,20,20),
tumor=c(50,70,40,60,80,90,40,50,70,80,50,70,40,60,80,90,40,50,70,80)
);
# 数据框重塑,数据合并为一列,添加分类列
d2=melt(d1,
variable.name="type",#新变量的名字
value.name="value" #值得名字
);
d2
######## 开始画图1 箱线图 + 散点图 done
ggplot(d2,aes(factor(type), value))+
geom_boxplot()+
geom_jitter()
######## 开始画图2 带误差bar的柱状图 + 散点图 done
#http://www.cookbook-r.com/Manipulating_data/Summarizing_data/
## Summarizes data.
## Gives count, mean, standard deviation, standard error of the mean, and confidence interval (default 95%).
## data: a data frame.
## measurevar: the name of a column that contains the variable to be summariezed
## groupvars: a vector containing names of columns that contain grouping variables
## na.rm: a boolean that indicates whether to ignore NA's
## conf.interval: the percent range of the confidence interval (default is 95%)
summarySE = function(data=NULL, measurevar, groupvars=NULL, na.rm=FALSE,
conf.interval=.95, .drop=TRUE) {
library(plyr)
# New version of length which can handle NA's: if na.rm==T, don't count them
length2 = function (x, na.rm=FALSE) {
if (na.rm) sum(!is.na(x))
else length(x)
}
# This does the summary. For each group's data frame, return a vector with
# N, mean, and sd
datac = ddply(data, groupvars, .drop=.drop,
.fun = function(xx, col) {
c(N = length2(xx[[col]], na.rm=na.rm),
mean = mean (xx[[col]], na.rm=na.rm),
sd = sd (xx[[col]], na.rm=na.rm)
)
},
measurevar
)
# Rename the "mean" column
datac = rename(datac, c("mean" = measurevar))
datac$se = datac$sd / sqrt(datac$N) # Calculate standard error of the mean
# Confidence interval multiplier for standard error
# Calculate t-statistic for confidence interval:
# e.g., if conf.interval is .95, use .975 (above/below), and use df=N-1
ciMult = qt(conf.interval/2 + .5, datac$N-1)
datac$ci = datac$se * ciMult
return(datac)
}
# http://www.cookbook-r.com/Graphs/Plotting_means_and_error_bars_(ggplot2)/
d3 = summarySE(d2, measurevar="value", groupvars=c("type"))
d3
ggplot(d3, aes(x=type, y=value)) +
geom_bar(aes(fill=type),position=position_dodge(), stat="identity", width=0.5) +
geom_errorbar(aes(ymin=value-ci, ymax=value+ci),
width=.2, # Width of the error bars
position=position_dodge(.9))+
geom_jitter(data=d2,aes(type,value), width=0.15) +
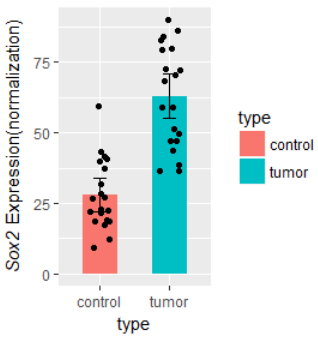
ylab( expression(paste( italic("Sox2")," Expression(normalization)") ) )
#ylab("Sox2 Expression\n(normalization)")
# V2.0 去掉图中文字的red/green glow
#怎么处理线性拟合r和p值
#1.1造数据
#x=data.frame(
# a=c(1,2,3,4,5),
# b=c(1,2,4,5,6)
#)
#1.2 抽样生成数据
library(dplyr)
set.seed(1)
sdata=sample_n(mtcars,100,replace=T)
x=data.frame(
a=sdata$mpg,
b=sdata$disp,
clazz=sdata$gear #分类变量
)
#方法1:用包计算r和p
#library(Hmisc)
#rs=rcorr(x$a,x$b, type="pearson")
#rs;str(rs)
#r=rs$r[2];r #[1] -0.8427099
#p=rs$P[2];p #[1] 0
#r0=round(r,2);r0 #[1] -0.84
#
#方法2:基础统计命令,计算r和p
#cor(x$a,x$b) #[1] -0.8427099
ts=cor.test(x$a,x$b); ts
str(ts)
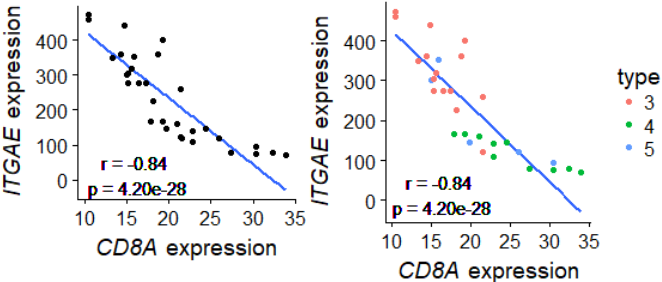
p=ts$p.value;p #[1] 4.202888e-28
r=ts$estimate[['cor']];r#[1] -0.8427099
r0=round(r,2);r0
# 保留两位有效数字
#https://stackoverflow.com/questions/39623636/forcing-r-output-to-be-scientific-notation-with-at-most-two-decimals
p0=formatC(p, format = "e", digits = 2)
p0
#可视化结果
#plot(x$a,x$b)
library(ggplot2)
library(cowplot)
g=ggplot(data=x,aes(a,b))+
geom_smooth(method='lm', se=F)+ #se=F不要置信区间的阴影
geom_text(data=data.frame(), aes(x=16,y=15,label=paste0("r = ",r0,"\n p = ",p0)))+
theme_cowplot() + #除掉主题背景阴影
xlab( expression(paste( italic("CD8A")," expression") ) )+
ylab( expression(paste( italic("ITGAE")," expression") ) )
g+geom_point() #一般图
g+geom_point(aes(color=factor(clazz)))+ #使用分类变量后
scale_color_discrete("type")
#目的: 给出数据矩阵,计算gene1和其余gene的相关系数,并画cor的heatmap。
# v0.1
#pheatmap 帮助文档 https://www.jianshu.com/p/1c55ea64ff3f
source("https://bioconductor.org/biocLite.R")
#biocLite("WGCNA")
#biocLite("ggplot2")
#biocLite("dplyr")
#biocLite("Seurat")
#biocLite("monocle")
setwd("C:\\Users\\DELL\\Desktop")
################
#read file H
msiH=read.csv("MSI-h.csv",row.names = 1)
msiH[1:3,1:3]
dim(msiH) #[1] 45 135
################
#read file
msiM=read.csv("MSIL-MSS.csv",row.names = 1)
msiM[1:3,1:3]
dim(msiM) #[1] 199 135
#check 重复项
genelist=colnames(msiM);genelist
#################
#defin function
getCorDF=function(msi){
#cor.test
ido1=msi$IDO1
df=NULL
for(i in 3:ncol(msi)){
data2=msi[,i]
symbol=colnames(msi)[i]
rs=cor.test(ido1, data2)
p=rs$p.value
correlation=as.numeric(rs$estimate)
tmp_df=data.frame(
symbol=symbol,
p=p,
correlation=correlation
)
#
df=rbind(df, tmp_df)
}
return(df)
}
df.h=getCorDF(msiH)
dim(df.h) #[1] 133 2
head(df.h)
row.names(df.h)=df.h$symbol
#df.h=df.h[,-1]
df.m=getCorDF(msiM)
dim(df.m) #[1] 133 2
row.names(df.m)=df.m$symbol
#df.m=df.m[,-1]
head(df.m)
cDF=data.frame(
symbol=row.names(df.m),
"MSI_H"=df.h$correlation,
"NON_MSI_H"=df.m$correlation,
row.names = 1
)
head(cDF)
dim(cDF) #[1] 133 2
########################
#heatmap
########################
library(pheatmap)
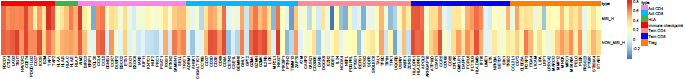
# 构建列 注释信息
type=read.csv("type.csv")
head(type)
dim(type) #[1] 133 2
annotation_col = data.frame(
#CellType = factor(rep(c("CT1", "CT2","CT3", "CT4","CT5"), 27))
type=type$type
)
rownames(annotation_col) = rownames(cDF) #type$sample # paste("Test", 1:10, sep = "")
head(annotation_col)
#
# 自定注释信息的颜色列表
ann_colors = list(
#Time = c("white", "firebrick"),
type = c("Act CD4" = "#FF81F2", "Act CD8" = "#00B9FF","HLA" = "#39B54A",
"immune checkpoint" = "#FF0000","Tem CD4" = "#FF8B99","Tem CD8" = "#0000FF",
"Treg" = "#FF8000")
)
head(ann_colors)
# pheatmap
library(Cairo)
CairoPDF("sunjiaxin_10_col.pdf",width=25,height=3)
pheatmap(t(cDF), cluster_rows=F,#是否聚类 row
cluster_cols=T, #是否聚类 列
#display_numbers = TRUE,number_color = "blue", #标上p值
annotation_col = annotation_col, #列 注释
annotation_colors = ann_colors,
angle_col=90,fontsize=10, #角度,字号
border=FALSE #没有边框
)
dev.off()
# V2.0 修改背景颜色为白色
#http://agetouch.blog.163.com/blog/static/22853509020161194123526/
# https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1323
# Using ggplot2 for volcano plots 使用ggplot2画火山图
library(ggplot2)
#读取数据 #data download from GEO2R result
setwd("C:\\Users\\Administrator\\Desktop")
dif=read.table(file="Primary Tumor_Normal Colon.txt",header=T,row.names=1)
dif[1:3,1:4]
#添加显著与否标签
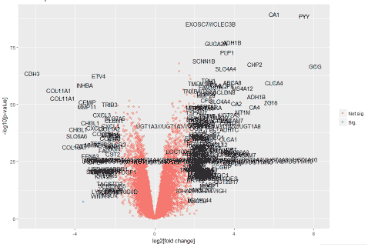
no_of_genes=nrow(dif);no_of_genes #4653
dif$threshold = as.factor(abs(dif$logFC) > 2 & dif$P.Value < 0.05/no_of_genes)
#画火山图
g = ggplot(data=dif, aes(x=logFC, y=-log10(P.Value), colour=threshold)) +
geom_point(alpha=0.4, size=1.75) +
#opts(legend.position = "none") +
theme(legend.title=element_blank()) +
scale_colour_hue(labels=c("Not sig.","Sig."))+
#xlim(c(-10, 10)) + ylim(c(0, 15)) +
xlab("log2[fold change]") + ylab("-log10[p-value]") +
theme_bw()+ # 背景色淡化
labs(title="Volcano plot")
g
#只标注显著基因的基因名
# 选出一部分基因:FC大且p小的基因
dd_text = dif[(abs(dif$logFC) > 2) & (dif$P.Value < 0.05/no_of_genes),]
head(dd_text)
#添加文字-基因名
g + geom_text(data=dd_text, aes(x=logFC, y=-log10(P.Value), label=dd_text$Gene.symbol), colour="black")
#为了防止遮挡,建议使用包添加文字
library(ggrepel)
g + geom_text_repel(data=dd_text, aes(x=logFC, y=-log10(P.Value), label=dd_text$Gene.symbol), colour="black")
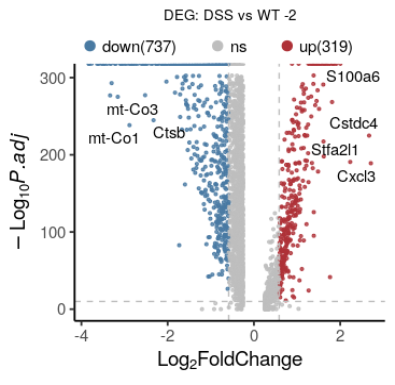
ref: https://blog.csdn.net/wangjunliang/article/details/123093894
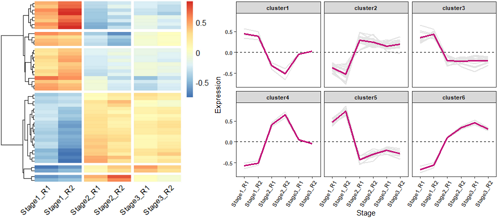
# 加载所需的R包
library(ggplot2)
library(pheatmap)
library(reshape2)
# 读取测试数据
Input =("
Stage1_R1 Stage1_R2 Stage2_R1 Stage2_R2 Stage3_R1 Stage3_R2
Unigene0001 -1.1777172 -1.036102 0.8423829 1.3458754 0.1080678 -0.08250721
Unigene0002 1.0596877 1.490939 -0.7663244 -0.6255567 -0.5333080 -0.62543728
Unigene0003 0.9206594 1.575844 -0.7861697 -0.3860003 -0.5501094 -0.77422398
Unigene0004 -1.3553173 -1.145970 0.2097526 0.7059886 0.9516353 0.63391053
Unigene0005 1.0134516 1.445897 -0.9705129 -0.8560422 -0.2556562 -0.37713783
Unigene0006 0.8675939 1.575735 -1.0120718 -0.5856459 -0.2821991 -0.56341216
")
data = read.table(textConnection(Input), header=TRUE,row.names=1)
##
#因为数据少,所以随便倍增一些数据。真实数据请跳过这一步
data=rbind(data,data*0.8) #造数据
data=rbind(data*0.9,data*(-0.8) ) #造数据
data=rbind(data*(-0.4),data*0.6) #造数据
row.names(data)=paste0('Unigene000',seq(1:nrow(data)) )#造数据
#end
#
#data = read.table("test.txt",header = T, row.names = 1,check.names = F)
# 查看数据基本信息
head(data)#View(data)
# 使用pheatmap绘制基因表达热图,并进行层次聚类分成不同的cluster
p = pheatmap(data, border=F, #不要描边
show_rownames = F, #不显示基因名
#cellwidth =40, #设置宽度
#scale ='row', #对每一行z标准化
cutree_rows = 6, #对row聚类时,设置加几个白横分割线
cluster_cols = F, #不对列聚类
gaps_col = c(2,4,6), #对列分割,仅用于不对列聚类的时候
angle_col = 45, #底部的字旋转的方向
fontsize = 12)
# 获取聚类后的基因顺序
row_cluster = cutree(p$tree_row,k=6)
# 对聚类后的数据进行重新排序
newOrder = data[p$tree_row$order,]
newOrder[,ncol(newOrder)+1]= row_cluster[match(rownames(newOrder),names(row_cluster))]
colnames(newOrder)[ncol(newOrder)]="Cluster"
# 查看重新排序后的数据
head(newOrder)
# 查看聚类后cluster的基本信息
unique(newOrder$Cluster)
#[1] 5 1 3 2 6 4
#统计每个cluster几个基因
table(newOrder$Cluster)
#1 2 3 4 5 6
#4 20 12 2 8 2
#
# 将新排序后的数据保存输出
newOrder$Cluster = paste0("cluster",newOrder$Cluster)
#write.table(newOrder, "expr_DE.pheatmap.cluster.txt",sep="\t",quote = F,row.names = T,col.names = T)
#
# 绘制每个cluster的基因聚类趋势图
newOrder$gene = rownames(newOrder)
head(newOrder)
# Stage1_R1 Stage1_R2 Stage2_R1 Stage2_R2 Stage3_R1 Stage3_R2 Cluster gene
#Unigene00032 0.4577851 0.6440856 -0.3310521 -0.2702405 -0.2303891 -0.2701889 cluster5 Unigene00032
#Unigene00033 0.3977249 0.6807646 -0.3396253 -0.1667521 -0.2376473 -0.3344648 cluster5 Unigene00033
#
#
library(reshape2)
# 将短数据格式转换为长数据格式
data_new = melt(newOrder)
head(data_new)
# Cluster gene variable value
#1 cluster5 Unigene00032 Stage1_R1 0.4577851
#2 cluster5 Unigene00033 Stage1_R1 0.3977249
# 绘制基因表达趋势折线图
ggplot(data_new,aes(variable, value, group=gene)) + geom_line(color="gray90",size=0.8) +
geom_hline(yintercept =0,linetype=2) +
stat_summary(aes(group=1),fun.y=mean, geom="line", size=1.2, color="#c51b7d") +
facet_wrap(Cluster~.) +
labs(x="Stage", y='Expression')+
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(size=8, face = "bold"),
axis.text.x=element_text(angle=60, hjust=1), #x标签旋转60度
strip.text = element_text(size = 8, face = "bold"))
#
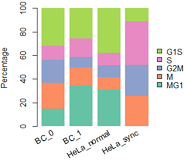
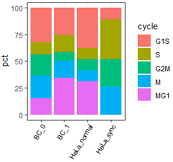
左: R原生画图; 右: ggplot2;
##data: 每群细胞中,各周期细胞数
mydatatxt="
G1S S G2M M MG1
BC_0 28 10 17 19 13
BC_1 21 13 7 13 28
HeLa_normal 11 3 3 3 9
HeLa_sync 3 10 7 7 0
"
#从字符串中读取数据框
tbl1=read.table(text=mydatatxt, header=T) # text设置了,file就要留空
tbl1=tbl1[,c(5,4,3,2,1)] #reorder columns
tbl1
#percentage
tbl2=apply(tbl1,1,function(x){ 100*x/sum(x)})
cellNames=colnames(tbl2)
colnames(tbl2)=NULL
tbl2
#画条状图
library(RColorBrewer)
col=RColorBrewer::brewer.pal(n = 5,name = "Set2");
col=rev(col)
barplot(1:5,col=col)
#plot
par(mar=c(5, 4, 4, 5) + 0.1)
posX=barplot(as.matrix(tbl2), col=col,
#names.arg=(paste(substr(FirstName,1,1),".",LastName)), #设定横坐标名称
names.arg=NULL,
space=0.2, #条形间距
#xlab="Individual #",
ylab="Percentage",
legend.text = rownames(tbl2),
args.legend=list(x="right", #border=NA, #不要图例小方块描边
box.col="white", inset=-0.25,bty="n"),
border=NA)
#加底部x坐标标签
y = -0.05;
text(posX, -5, labels=cellNames, adj=1, srt=30, xpd=TRUE) #adj标签与轴的距离,srt设置xlable角度
#box()
## 使用ggplot2的版本。
mydatatxt="
G1S S G2M M MG1
BC_0 28 10 17 19 13
BC_1 21 13 7 13 28
HeLa_normal 11 3 3 3 9
HeLa_sync 3 10 7 7 0
"
#从字符串中读取数据框
tbl1=read.table(text=mydatatxt, header=T) # text设置了,file就要留空
#转为百分数
tbl2=as.data.frame(apply(tbl1, 1, function(x){x/sum(x)*100}) )
tbl2$cycle=rownames(tbl2) #添加一列:行名
tbl2
# BC_0 BC_1 HeLa_normal HeLa_sync cycle
#G1S 32.18391 25.609756 37.93103 11.11111 G1S
# 变为一列
dt2=reshape2::melt(tbl2,
id.vars='cycle', #要保留的id变量(源)
variable.name = "type", #melt后变量名列的名字(目标)
#measure.vars=c(), #要melt的变量名(列名)(源)
value.name="pct"); #melt后的数据列名字(目标)
head(dt2)
# cycle type pct
#1 G1S BC_0 32.18391
#定义图例顺序
dt2$cycle=factor(dt2$cycle, levels=c('G1S','S','G2M','M','MG1'))
ggplot(dt2, aes(x=type,y=pct, fill=cycle ))+
geom_bar(stat='identity')+
theme_bw()+
#+theme(legend.position = "none")+ # 不显示坐标轴
labs(x="")+
theme(axis.text.x=element_text(angle=60, hjust=1,size=8,color="black") ) #坐标轴文字旋转60度
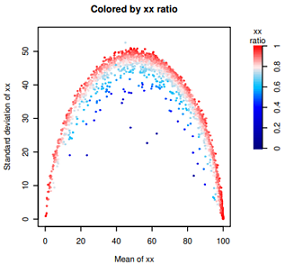
数据特点:范围是0-1,但是0.5以下很稀疏,峰值在0.8-0.9之间,1也是一个峰。
由于颜色变量偏斜太厉害,直接使用ggplot2,渐变色只能指定3种颜色,偏斜导致整体颜色太淡。
目前不会使用ggplot2设置4种颜色的渐变色,只好手动用R原生函数绘制了。
head(dif)
# gene cellNumber meanDPAU sdDPAU RNACounts cpm totalCell domCell nonDomCell ratio
#RPL8 RPL8 225 99.86553 0.3462266 712259 492539.3 225 225 0 1.0000000
#RPL3 RPL3 225 99.73653 0.9470744 542531 369992.2 225 223 2 0.9911111
#...
#fig1: 查看分布图,略。
hist(dif$ratio, n=200)
abline(v=0.9, col='red', lty=2)
#
#fig2: 略。(点的颜色普遍太淡)
library(ggplot2)
ggplot(dif, aes(meanDPAU, sdDPAU, color=ratio))+
geom_point(size=0.2)+
scale_color_gradient2('xx\nRatio', low="navy", mid='white', high="red", midpoint =0.7)+
theme_bw()
#fig3: 见上图
# step0: 设置颜色。色彩向量取值范围是0-1,但是数据偏斜向1。0.5以下很稀疏。
bk0=seq(0.0,0.39,by=0.01) #almost non data
bk1=seq(0.4,0.59,by=0.01)
bk2=seq(0.6,0.79,by=0.01)
bk3=seq(0.8,1,by=0.01)
#
colors = c(
colorRampPalette(colors = c("navy","blue"))(length(bk0)),
colorRampPalette(colors = c("blue","#00BFFF"))(length(bk1)),
colorRampPalette(colors = c("#00BFFF","#FFE9E9"))(length(bk2)),
colorRampPalette(colors = c("#FFE9E9","red"))(length(bk3))
)
print( length( seq(0,1,0.01) ) )
print( length(colors) )
#
library(Cairo)
dev.off() #for jupyter bug
CairoPDF('6_9_domi__testing__.pdf', width=4.5,height=4)
#页面布局
mat = matrix(c(1,1,1,1,1,1,2,2), nrow=1, byrow=TRUE);
layout(mat)
############
#step1: plot
par(mar=c(4.5,4.5,4,0)) #bottom, left, top, right
plot(NULL, xlim=c(0,100), ylim=c(0,55), type='n',
xlab="Mean of xx",
ylab="Standard deviation of xx",
main="Colored by xx ratio")
for(i in 1:nrow(dif)){
v=round(dif$ratio[i],2)*100
color=colors[v]
points(dif[i,'meanDPAU'], dif[i, 'sdDPAU'],
pch=20, cex=0.05, col=color)
}
############
#step2: 用image函数画color bar
par(mar=c(13,2.5,6,5)) #bottom, left, top, right
n=100
image(t(matrix(0:n)),col=colors,
xaxt="n", yaxt="n", cex = 1.5,
mgp=c(0,1,0),
#main="Dominant\ncell ratio",
bty='n',#box type
xlab = "", ylab = ""
)
text(x=0,y=1.1,labels=c("xx\nratio"),xpd=T)
axis(4,at=seq(0,1,by=0.2),
labels=seq(0,1,0.20),
cex.axis=1, #坐标轴刻度字体大小
#col.axis="red", #坐标轴刻度字体颜色
#col="purple", #坐标轴颜色
lwd=1, #坐标轴刻度粗细
las=0)#las=0垂直于轴,2平行于轴
#
dev.off()
#cut(x, breaks): Convert Numeric to Factor
# 1. 生成数据
set.seed(20211119)
dat_1=rnorm(100, 0, 1)
# 3. 数据和颜色的映射
# fun1: 使用cut 函数,连续型变量变成分窗的因子
continuousDat2bins=function(dat, binwidth=NA){
# to get min and max
l1=min(dat);l1 #-1.868331
u1=max(dat);u1 #2.796254
l2=floor(l1*10)/10; l2 #-1.9
u2=ceiling(u1*10)/10; u2 #2.8
# get binwidth
if(is.na(binwidth)){
binwidth=(u2-l2)/100
}
# get breaks, include the max number
breaks=seq(l2, u2, binwidth)
if(max(breaks) < u2){ breaks=c(breaks, u2)}
# convert dat to factors
dat_3=cut(dat, breaks = breaks );
dat_3
}
#dat_3=continuousDat2bins(dat_1)
# fun2: 分窗的因子变为渐变色颜色
bins2colors=function(bins, colors=c('green4','white','maroon3')){
# 定义渐变色
colors2=colorRampPalette(colors = colors, interpolate ="linear")( length(levels(bins)) )
# map dat to colors, by index of bin index
plotcol = colors2[ bins ];
return(list(plotcol, colors2) )
}
#bins2colors(dat_3)
# fun3: 合并以上2个函数,连续型变量映射到渐变色颜色
continuousDat2Colos=function(dat, binwidth=NA, colors=c('green4','white','maroon3')){
bins2colors(continuousDat2bins(dat, binwidth), colors )
}
#continuousDat2Colos(dat_1)
colors2=continuousDat2Colos(dat_1, colors=c("purple", "purple4", "black", "yellow3", "yellow"))
colors2
# 4. 画图
#pdf("01.pdf", width=3.2, height=2.8)
png("01.png", width=3.2*72, height=2.8*72, res=72)
mat=matrix(c(1,2),ncol=2)
layout(mat, widths = c(3,1)) #右图: 左右3:1
par(mar=c(3,3,1,0))
# left
plot(dat_1, pch=20,
col=colors2[[1]],
bty="l",
mgp=c(2,0.8,0),
ylab="XX value",
main="")
box(bty="l", lwd=2) #坐标轴加粗
# right
#par(mar=c(3,1,3,5))
par(mar=c(7,0.5,2, 3))
len=length(colors2[[2]]) #颜色长度
barplot( rep(1, len), col=colors2[[2]],
border=NA, space=0, horiz =T,
ann=F, axes=F)
# 图例上的数字怎么能靠近整数关口? //todo
# fun: 图例表示的数值data,渐变色的长度len,获取渐变色上的刻度位置和刻度值。
getTicks=function(dat, len){ #需要手动修正
# (Real)from min(dat) to max(dat)
# (mark)to 0 to len
real_tick_len=(max(dat) - min(dat)) / 4
real_tick=seq(min(dat), max(dat), real_tick_len)
real_tick=round(real_tick, 2)
mark_tick=seq(0, len, (len-0)/4)
return( list(
label=real_tick, #等差数列,来自dat,刻度值
at=mark_tick # 等差数列,来自渐变色长度 len,刻度位置
) )
}
ticks=getTicks(dat_1, len)
axis(side=4, at=ticks$at, #刻度位置
labels=ticks$label, #刻度值
tcl=-0.1, # 刻度线长度
mgp=c(1,0.3,0), #刻度值与颜色条距离
las=2)
dev.off()
组合图,比如要求共用x坐标轴时,很难用ggplot2来处理,这时候可以考虑原生绘图,相当给力!
示例: 顶部barplot,底下百分数barplot,共用x坐标轴,所以要一一对应。
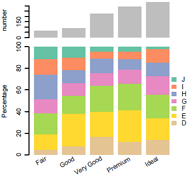
library(Cairo)
# make data
dt0=diamonds[1:1000,]
head(dt0)
dt1=table(dt0$cut, dt0$color)
dt1
CairoPDF('barplot_combine.pdf', width=4,height=4) #要在布局代码之前保存
#页面布局
mat = matrix(c(1,1,2,2,2,2), ncol=1, byrow=T);mat
layout(mat)
### fig1 barplot
dt.rsum=apply(dt1, 1, sum)
dt.rsum
par(mar=c(1,4,4,6)+ 0.1) #bottom, left, top, right
barplot(as.numeric(dt.rsum), ylab="number",
names.arg=NULL,border=NA,space=0.2)
#### fig2 percentage barplot
tbl2=apply(dt1,1,function(x){ 100*x/sum(x)})
cellNames=colnames(tbl2)
colnames(tbl2)=NULL
tbl2
#画条状图
library(RColorBrewer)
col=RColorBrewer::brewer.pal(n = nrow(tbl2),name = "Set2");
col=rev(col)
#barplot(1:5,col=col)
#plot
par(mar=c(5, 4, 0, 6) + 0.1) ##bottom, left, top, right
posX2=barplot(as.matrix(tbl2), col=col,
#names.arg=(paste(substr(FirstName,1,1),".",LastName)), #设定横坐标名称
names.arg=NULL,
space=0.2, #条形间距
#xlab="Individual #",
ylab="Percentage",
legend.text = rownames(tbl2),
args.legend=list(x="right", border=NA, #不要图例小方块描边
box.col="black", inset=-0.1,bty="n"),
border=NA)
# 加底部x坐标标签
y2 = par('usr')[3]-2;
text(posX2, y2, labels=cellNames, adj=1, srt=30, xpd=TRUE) #adj标签与轴的距离,srt设置xlable角度
#box()
dev.off()
## 绘图区域的大小
# par('usr') #c(x1, x2, y1, y2)
#[1] -0.032 6.232 -1.000 100.000
#附: fig1 barplot 如果想在bar顶部标记上数字和百分比
dt.rsum=apply(dt1, 1, sum);dt.rsum
dt.rsum=as.numeric(dt.rsum);dt.rsum
par(mar=c(1,4,5,6)+ 0.1) #bottom, left, top, right
posX=barplot(dt.rsum, ylab="number",
ylim=c(0,400),
names.arg=NULL,border=NA,space=0.2)
text(x=posX, y=dt.rsum+20, label=paste0(dt.rsum, '\n(', round(dt.rsum/sum(dt.rsum),2)*100,'%)' ),
font=1,cex=0.7)
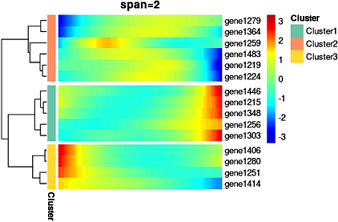
感觉调试的影响有可能大于生物学意义本身,只能看大趋势,细节的起伏很大程度上被平滑化掩盖掉了。
1.对于细胞,按照monocle等伪时间顺序排列。
2.对于基因,使用了log2(cpm+1)作为表达量,按照表达量最高的前20%细胞的中位数排序,选前几十个基因。
3.对每个基因,使用 LOESS 做平滑化后的基因表达值做heatmap。
对本模拟数据而言,span=1左右平滑化似乎最优。
目标图片 monocle at github:
分化过程图 | 2个分化方向 |
下面是本代码效果图。
#调试和探索函数
check=function(df){
print(dim(df))
print(head(df))
}
# 负二项分布,模拟基因的表达,比正态分布更优。
a1=c(); a2=c()
for(i in 1:1000){
w=rnbinom(cellNumber, size = 100, prob = 0.1)
#plot(w)
a1=c(a1, mean(w) )
a2=c(a2, sd(w))
}
plot(a1,a2)
w=rnbinom(cellNumber, size = 100, prob = 0.1)
hist(w)
############### color bar from monocle::plot_pseudotime_heatmap ##############
# 这个 color bar 来自于monocle,画热图效果很好。该渐变色原理和细节请参考 http://www.biomooc.com/R/R-color.html#2_3
# fn1
table.ramp = function (n, mid = 0.5, sill = 0.5, base = 1, height = 1) {
x <- seq(0, 1, length.out = n)
y <- rep(0, length(x))
sill.min <- max(c(1, round((n - 1) * (mid - sill/2)) + 1))
sill.max <- min(c(n, round((n - 1) * (mid + sill/2)) + 1))
y[sill.min:sill.max] <- 1
base.min <- round((n - 1) * (mid - base/2)) + 1
base.max <- round((n - 1) * (mid + base/2)) + 1
xi <- base.min:sill.min
yi <- seq(0, 1, length.out = length(xi))
i <- which(xi > 0 & xi <= n)
y[xi[i]] <- yi[i]
xi <- sill.max:base.max
yi <- seq(1, 0, length.out = length(xi))
i <- which(xi > 0 & xi <= n)
y[xi[i]] <- yi[i]
height * y
}
# fn2
rgb.tables=function (n, red = c(0.75, 0.25, 1), green = c(0.5, 0.25, 1), blue = c(0.25, 0.25, 1)) {
rr <- do.call("table.ramp", as.list(c(n, red)))
gr <- do.call("table.ramp", as.list(c(n, green)))
br <- do.call("table.ramp", as.list(c(n, blue)))
rgb(rr, gr, br)
}
# fn3
blue2green2red=function (n) {
rgb.tables(n, red = c(0.8, 0.2, 1), green = c(0.5, 0.4, 0.8), blue = c(0.2, 0.2, 1))
}
bks <- seq(-3.1,3.1, by = 0.1)
hmcols <- blue2green2red(length(bks) - 1)
hmcols
# view the color bar
barplot(rep(1, length(hmcols)), col=hmcols, border = NA, space=0, axes=F)
############### make data: 造数据,如果时真实数据,跳过这一步 ##############
makeData=function(){
df=NULL
set.seed(2020)
for(i in 1:geneNumber){
#a=rnorm(cellNumber, mean = 3, sd = 5)
a=rnbinom(cellNumber, size = 1000, prob = 0.1) #负二项分布
names(a)=paste0('cell', 1:cellNumber)
df=rbind(df, a)
}
#rownames(df)=paste0('gene', 1:geneNumber)
dim(df)
df[1:10,1:5]
## 倍增仅
df=rbind(df,df*0.8) #造数据
df=rbind(df*1.9,df*(-1.8) ) #造数据
df=rbind(df*(-0.4),df*0.6) #造数据
#
#负值变正数
df=apply(df, 2, function(x){
ifelse(x<0, -10*x, 10*x)
})
df=as.data.frame(df)
# 要去掉全是0的行
keep=apply(df,1,sum)>0
df=df[keep,]
row.names(df)=paste0('gene',seq(1:nrow(df)) )#造数据
return(df)
}
#
cellNumber=100
geneNumber=300
df2=makeData()
dim(df2)
df2[1:10,1:5]
# normalization
df2.cpm=apply(df2, 2, function(x){1e6*x/sum(x)})
df2.log2cpm=apply(df2.cpm, 2, function(x){log2(x+1)})
dim(df2.log2cpm)
head(df2.log2cpm[,1:4])
library(pheatmap)
pheatmap( df2, scale='row')
pheatmap( df2.log2cpm, scale='row') #这热图没法看
###
## step2: select genes, by expression
#筛选前10%细胞表达median值最高的10%的基因
median_df=NULL
i=1
for(gene in rownames(df2.log2cpm)){
i=i+1
#if(i>10)break;
tmp=as.numeric(df2.log2cpm[gene,])
tmp=tmp[order(-tmp)]
tmp.median=median(tmp[1: round(length(tmp)*0.1)])
median_df=rbind(median_df, data.frame(
gene=gene,
value=tmp.median,
sd=sd(tmp)
))
}
rownames(median_df)=median_df$gene
median_df=median_df[order(-median_df$value),]
check(median_df)
useGenes=rownames(median_df)[1:round(geneNumber*0.05)]
length(useGenes)
head(useGenes)
# 使用标准化后的数据
p=pheatmap(df2.log2cpm[useGenes,] , border_color = NA, scale='row',
color=hmcols, main="log2cpm")
#使用原始counts数据
#p=pheatmap(df2[useGenes,] , border_color = NA, scale='row',
# color=hmcols, main="raw counts")
#get cell order from heatmap, and reorder df
cellOrder=colnames(df2.log2cpm)[p$tree_col$order]
head(cellOrder)
df3=df2.log2cpm[,cellOrder]
head(df3[,1:4])
########################
## LOESS 平滑化
########################
##########
# test one gene
geneID=1
y1=df3[geneID,]
plot(y1, type='l')
#
addLine=function(span, color, ...){
y2=predict(loess(df3[geneID,] ~ seq(1, ncol(df3)), span=span))
lines(y2, type="l", col=color, ...)
}
addLine(0.1, 'green')
addLine(0.2, 'blue')
addLine(0.5, 'red') #差不多了
addLine(1, 'purple',lwd=2)
addLine(2, 'orange',lwd=2, lty=2)
addLine(8, 'grey',lwd=5, lty=2)
##########
## 批量化
testSpan=function(span=0.2){
df4=apply(df3, 1, function(x){
predict(loess(x ~ seq(1, length(x)), span=span))
})
df4=as.data.frame(t(df4))
colnames(df4)=colnames(df3)
dim(df3)
dim(df4)
head(df4[,1:5])
## heatmap again
p=pheatmap(df4[useGenes,] , border_color = NA, scale='row',
clustering_method='ward.D2',
cluster_cols = F,
show_colnames = F,
#gaps_col = c(60),
cutree_rows = 3,
color=hmcols, main=paste0("span=", span) )
return(p)
}
# test various span for perfect effect
testSpan(0.1)
testSpan(0.5)
testSpan(1) #效果可以
testSpan(1.5) #
testSpan(2) #效果可以
testSpan(4)
testSpan(8) #之后无变化
testSpan(16)
testSpan(32)
testSpan(100)
#
testSpan(0.2)
testSpan(0.3)
testSpan(0.4)
testSpan(0.5)#
testSpan(0.6)
testSpan(0.7)
testSpan(0.8)
testSpan(0.9)
# 重绘热图,add row annotation
p2=testSpan(2)
# 获取聚类后的基因顺序
row_cluster = cutree(p2$tree_row,k=3)
row_cluster
# 获取每个cluster的基因名字
gSet1=names(row_cluster[which(row_cluster==1)]) #示例
#
annote_row=data.frame(
gene=names(row_cluster),
Cluster=paste0("Cluster",unname(row_cluster)),
row.names = 1
)
annote_row$Cluster=as.factor(annote_row$Cluster)
head(annote_row)
## 这个自定义颜色必须是list,不能是df!!
annote_color=list(
Cluster=c('Cluster1'="#66C3A6", 'Cluster2'="#FD8D63",'Cluster3'="#FFD92F")
)
head(annote_row)
testSpan2=function(annote_row,annote_color, span=0.2){
df4=apply(df3, 1, function(x){
predict(loess(x ~ seq(1, length(x)), span=span))
})
df4=as.data.frame(t(df4))
colnames(df4)=colnames(df3)
dim(df3)
dim(df4)
head(df4[,1:5])
## heatmap again
p=pheatmap(df4[useGenes,] , border_color = NA, scale='row',
clustering_method='ward.D2',
cluster_cols = F,
show_colnames = F,
annotation_row = annote_row,
annotation_colors = annote_color,
#gaps_col = c(60),
cutree_rows = 3,
color=hmcols, main=paste0("span=", span) )
return(p)
}
testSpan2(annote_row,annote_color, span=2) #页码上面的示例图就是这一步的结果
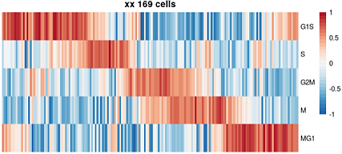
目标图片: 2015 Cell, Drop seq, Figure 4 Cell-Cycle Analysis of HEK and 3T3 Cells Analyzed by Drop-Seq.
细胞周期相关基因 list: A complete list of cell-cycle regulated genes can be found in Table S2
原文是一段非常高质量的R代码,充分展示了如何嵌套使用apply家族函数、如何函数嵌套、如何定义输入和输出。
下文为了方便理解,把原来的嵌套函数拆开了。

# aim: scRNAseq to determine cell cycle.
# 缺陷: 这是对细胞系做的周期分类,没有考虑G0期。
# source: 2015 cell, drop seq;
# v0.3 fix filter line.
## 准备工作
#1. 定义资源的路径: infoPath
infoPath="/home/wangjl/data/cellLines/"
#basic information, like cycle gene list, RNA expression matrix, cell type info;
#2.下载周期相关基因列表,命名为 G1S.txt等5个文件,一个基因一行,不加引号。文件放到infoPath下。
#3. 你自己的单细胞转录组表达矩阵, row为基因,column为cell;
#4. cell information: 可选参数,用于对表达矩阵的column做筛选,也就是取细胞的子集。
# 经验:
#1. 一次对一种细胞做细胞周期鉴定,超过一个种类可能结果不准确;
#2. 如果细胞的一部分是(药物、刺激等)处理过的,那么用normal部分筛选基因后,对全部细胞做周期鉴定,否则会失真。
#3. 要保证矩阵不能有全是0的行。本例是3'测序,使用的是log(cpm+1),如果是全长测序,可以使用log(tpm+1)或log(rpkm+1)
setwd("/home/wangjl/data/cellLines/cycle/")
outputRoot="BC_"
#load data: 表达counts矩阵 (确定是 raw counts矩阵,第一步过后会有标准化)
fname=paste0(infoPath,"BC_HeLa.225cells.count.V3.csv")
data=read.csv(fname,row.names = 1)
dim(data) #[1] 18679 225
data[1:4,1:4]
# c01ROW24 c01ROW35 c01ROW31 c05ROW02
#A1BG-AS1 0 0 0 0
#A2M 0 0 0 0
#helper: return cell phase names
getCellPhaseList=function(){
c('G1S', 'S','G2M','M','MG1');
}
#load data: 细胞类型
cellType=read.csv( paste0(infoPath,"cellInfo.txt"),row.names = 1)
head(cellType)
# geneNumber countsPerCell countsPerGene cellType
#c01ROW24 5693 1950601 342.6315 BC_syncMix
#c01ROW35 6768 3048970 450.4979 BC_syncMix
dim(cellType) #225 4
#获取细胞子集:BC
data2=data[,row.names(cellType[which( substr(cellType$cellType,1,3)=="BC_" ),])]
dim(data2) #[1] 18679 169
#只保留非0的行
keep=apply(data2>0,1,sum)>0
table(keep)
#FALSE TRUE
#403 18276
data2=data2[keep,] #filter out all 0 rows
dim(data2) #[1] 18276 169
#load data: 每个时期的gene set,和表达矩阵求基因交集
geneSets=list();
for(i in 1:length(getCellPhaseList() )){
setName=getCellPhaseList()[i]
print( paste(i, setName) )
tmpGenes=readLines( paste0(infoPath,setName,'.txt') )
print( length(tmpGenes) )
#和矩阵的列求交集
geneSets[[setName]]=intersect(tmpGenes, row.names(data2))
print( length(geneSets[[setName]])) #95
}
## begin
#step1: exclude genes cor<0.3 with mean of the set
geneSets2=list();
for(i in 1:length(getCellPhaseList() )){
#求G1S集合中的gene mean
setName=getCellPhaseList()[i] #'G1S';
print( paste(i, setName) )
setMean=apply(data2[geneSets[[setName]], ], 2,mean)
#head(setMean)
#每个基因和平均值求cor
tmpGenes=c()
for(g in geneSets[[setName]]){
#rs=cor(t(data2[g,]), setMean)
rs=cor(as.numeric(data2[g,]), setMean)
#print( paste(g,rs) )
if(rs>=0.3){
tmpGenes=c(tmpGenes,g)
}
}
#[1] "ACD 0.324866931495592"
#[1] "ACYP1 0.0858240480040331"
#[1] "ADAMTS1 -0.128818193750771"
print( length(tmpGenes) )
geneSets2[[setName]]=tmpGenes
}
geneSets2
sum(sapply(geneSets2, function(x){length(x)})) #321
# step2: depth norm; log2 norm;
getNormalizedCts <- function ( cts ) {
#ctsPath
#cts <- read.table ( ctsPath , header = T , as.is = T )
apply ( cts , 2 , function ( x ) { log2 ( ( 10^6 ) * x / sum ( x ) + 1 ) })
}
normCts=getNormalizedCts(data2)
dim(normCts)
normCts[1:10,1:5]
#apply(normCts,2,sum)
#step3: calculate 5 phase scores each cell(mean of phase genes)
# Tested. Passed.
assignSampleScore <- function ( phaseGenesList , normCts ) {
scores <- lapply ( phaseGenesList , function ( pGenes ) {
print(length(pGenes) )
apply ( normCts , 2 , function ( x ) {
mean ( x [ pGenes ] )
} )
} )
do.call ( cbind , scores )
}
scoreMatrix <- assignSampleScore ( geneSets2 , normCts )
dim(scoreMatrix) #169 5
head(scoreMatrix)
# G1S S G2M M MG1
#c12ROW02 3.774424 3.742809 3.131765 3.524596 4.595119
#c12ROW03 3.274414 2.999720 3.231767 3.422519 5.221042
write.table ( scoreMatrix , paste ( outputRoot , "PhaseScores.txt" , sep = "" ) )
# pheatmap(scoreMatrix, scale='row', border_color = NA)
#step4: z-norm(each phase, then each cell)
# Tested. Passed.
getNormalizedScores <- function ( scoreMatrix ) {
norm1 <- apply ( scoreMatrix , 2 , scale )
normScores <- t ( apply ( t ( norm1 ) , 2 , scale ) )
rownames ( normScores ) <- rownames ( scoreMatrix )
colnames ( normScores ) <- colnames ( scoreMatrix )
normScores
}
normScores <- getNormalizedScores ( scoreMatrix )
head(normScores)
# G1S S G2M M MG1
#c12ROW02 -0.5587886 1.71406144 -0.767616785 0.012780937 -0.4004370
#c12ROW03 -1.0458011 -0.56059069 -0.005651298 0.002073499 1.6099696
write.table ( normScores , paste ( outputRoot , "PhaseNormScores.txt", sep = "" ) )
#pheatmap(normScores, scale='row', border_color = NA)
#画出每个细胞的各个周期打分
i=5;plot(normScores[i,],type='o',col=rainbow(i), main=rownames(normScores)[i])
i=8;plot(normScores[i,],type='o',col=rainbow(i), main=rownames(normScores)[i])
#step5: assign phase for each cell
getReferenceProfiles <- function () {
referenceProfiles <- list (
"G1S" = c ( 1 , 0 , 0 , 0 , 0 ) ,
"G1S.S" = c ( 1 , 1 , 0 , 0 , 0 ) ,
"S" = c ( 0 , 1 , 0 , 0 , 0 ) ,
"S.G2M" = c ( 0 , 1 , 1 , 0 , 0 ) ,
"G2M" = c ( 0 , 0 , 1 , 0 , 0 ) ,
"G2M.M" = c ( 0 , 0 , 1 , 1 , 0 ) ,
"M" = c ( 0 , 0 , 0 , 1 , 0 ) ,
"M.MG1" = c ( 0 , 0 , 0 , 1 , 1 ) ,
"MG1" = c ( 0 , 0 , 0 , 0 , 1 ) ,
"MG1.G1S" = c ( 1 , 0 , 0 , 0 , 1 ) ,
"all" = c ( 1 , 1 , 1 , 1 , 1 ) )
#referenceProfiles <- lapply ( referenceProfiles , function ( x ) { names ( x ) <- c ( "G1S", "S", "G2" , "G2M" , "MG1" ); x } )
referenceProfiles <- lapply ( referenceProfiles , function ( x ) {
names ( x ) <- c ( 'G1S', 'S','G2M','M','MG1' ); x } )
do.call ( rbind , referenceProfiles )
}
# Tested. Passed.
assignRefCors <- function ( normScores ) {
referenceProfiles <- getReferenceProfiles()
t ( apply ( normScores , 1 , function ( sampleScores ) {
apply ( referenceProfiles , 1 , function ( refProfile ) {
cor ( sampleScores , refProfile ) } )
} ) )
}
# Tested. Passed.
getPhases <- function ( ) {
#phases <- c ( "G1S", "S", "G2" , "G2M" , "MG1" )
phases <- c ( 'G1S', 'S','G2M','M','MG1' )
names ( phases ) <- phases
phases
}
# Tested. Passed.
assignPhase <- function ( refCors ) {
phases <- getPhases ()
apply ( refCors [ ,phases ] , 1 , function ( x ) {
phases [ which.max ( x ) ]
} )
}
###### Score cycle similarity
refCors <- assignRefCors ( normScores )
head(refCors)
assignedPhase <- assignPhase ( refCors )
head(assignedPhase)
#
table(assignedPhase)
# assignedPhase
#G1S G2M M MG1 S
#49 24 32 41 23
#
getDFfromNamed=function(Namedxx){
data.frame(
id=attr(Namedxx,'names'),
val=unname(Namedxx),
row.names = 1
)
}
rs=getDFfromNamed(assignedPhase)
head(rs)
#保存标签结果
write.csv(rs,paste ( outputRoot , "cellCycle_phase.csv" , sep = "" ) )
#
#和第四步比较呢?
i=5;plot(normScores[i,],type='o',col=rainbow(i), main=rownames(normScores)[i])
#目测几个,就是求最高值
#
#step6 ####### Order cells
# Tested. Passed.
orderSamples <- function ( refCors , assignedPhase) {
phases <- getPhases()
orderedSamples <- list()
for ( phase in phases ) {
phaseCor <- refCors [ assignedPhase == phase , ]
phaseIndex <- which ( colnames ( phaseCor ) == phase )
if ( phaseIndex == 1 ) { preceding = ncol ( phaseCor ) - 1 } else { preceding <- phaseIndex - 1 }
if ( phaseIndex == ncol ( phaseCor ) - 1 ) { following = 1 } else { following <- phaseIndex + 1 }
earlyIndex <- phaseCor [ , preceding ] > phaseCor [ , following ]
earlyCor <- subset ( phaseCor , earlyIndex )
earlySamples <- rownames ( earlyCor ) [ order ( earlyCor [ , preceding ] , decreasing = T ) ]
lateCor <- subset ( phaseCor , ! earlyIndex )
lateSamples <- rownames ( lateCor ) [ order ( lateCor [ , following ] , decreasing = F ) ]
orderedSamples [[ phase ]] <- c ( earlySamples , lateSamples )
}
refCors [ do.call ( c , orderedSamples ) , ]
}
#
ordCor <- orderSamples ( refCors , assignedPhase )
write.table ( cbind ( ordCor , "assignedPhase" = assignedPhase [ rownames ( ordCor ) ] ) ,
paste ( outputRoot , "PhaseRefCor.txt" , sep = "" ) )
#
#step7 ####### Plot
# Passed.
plotCycle <- function ( phaseCorsMatrix, ... ) {
library("pheatmap")
library("RColorBrewer")
breaks <- seq ( -1 , 1 , length.out = 31 )
heatColors <- rev (brewer.pal ( 9, 'RdBu'))
heatColors <-colorRampPalette(heatColors)
colorPallete <- heatColors((length ( breaks ) - 1 ))
# create heatmap
hm.parameters <- list(phaseCorsMatrix, border_color=NA,
color = colorPallete,
breaks = breaks,
cellwidth = NA, cellheight = NA, scale = "none",
treeheight_row = 50,
kmeans_k = NA,
show_rownames = T, show_colnames = F,
#main = "",
clustering_method = "average",
cluster_rows = FALSE, cluster_cols = FALSE,
clustering_distance_rows = "euclidean",
clustering_distance_cols = NA ,
legend = T , annotation_legend = F,... )
do.call("pheatmap", hm.parameters )
}
phases <- getPhases()
#jpeg ( paste ( outputRoot , "PhasePlot.jpg" , sep = "" ) , 5 , 3 , units = "in" , res = 300 )
pdf( paste0( outputRoot, "a7_PhasePlot.pdf"), width=4, height=2.5)
plotCycle ( t ( ordCor [ , phases ] ), main= paste0("XX ", nrow(ordCor), ' cells') )
#dev.off()
## check
df1=rs
head(df1)
table(df1$val)
cellType2=cellType[rownames(df1),]
cellType2$cellCycle=df1$val
head(cellType2)
table(cellType2$cellType, cellType2$cellCycle)
# G1S G2M M MG1 S
#BC_0 28 17 19 13 10
#BC_1 21 7 13 28 13
目标图片: Science 2018, Fig. 5 The molecular clock across tissues
(C) Distribution of the peak phases of core clock components in the tissues where they are detected as cycling.
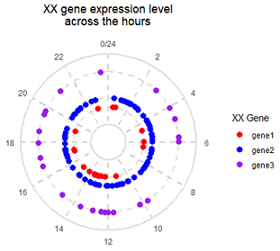
#1. make data
n=200
set.seed(10)
dt=data.frame(
time=runif(n, 0,24),
gene1=rbinom(n, 1, 0.1),
gene2=rbinom(n, 1, 0.4),
gene3=rbinom(n, 1, 0.1)
)
dt[which(dt$gene1==1),]$gene1=7
dt[which(dt$gene2==1),]$gene2=7.5
dt[which(dt$gene3==1),]$gene3=9
#table(dt$gene3)
library(reshape2)
df=melt(dt, id="time")
head(df)
# 第一列时间点(0-24), 第二列基因名字,第三列基因表达是与否(0或1)
# time variable value
#1 12.179477 gene1 0
#2 7.362444 gene1 0
#3 10.245784 gene1 0
#2. draw
draw=function(df){
#step1 定义一个无数据的空坐标系统
g=ggplot()+
theme_bw()+
theme(panel.grid =element_blank(), ## 删去网格线
panel.border = element_blank(), #去掉外边框
plot.title = element_text(hjust = 0.5), #标题居中
axis.text.y = element_blank(), #删掉y轴文字
axis.ticks = element_blank() #删去坐标刻度
) +
#coord_cartesian(ylim=c(1.5,2.8))+
scale_y_continuous( expand = c(0,0), #去掉图与y轴间隙
limits=c(5,10))+ #y轴显示范围
scale_x_continuous(expand = c(0,0),
breaks=seq(0,24,2), #每2个一个刻度
labels=seq(0,24,2))+
labs(x="", y="", title="XX gene expression level\nacross the hours")
#step2 添加网格线: 放射线
g=g + geom_vline(aes(xintercept= seq(0,24,2) ), color="#D0D0D0", size=0.8,
linetype="dashed")
#step3 添加网格线: 圆圈线
for(i in c( 7, 7.5, 9)){
g=(function(i2){
print(i2)# 不加这一句不更新,会遇到闭包问题
g + geom_hline(aes(yintercept=i2), color="#DFDFDF", size=1,
linetype="dashed")
})(i)
}
g=g + geom_hline(aes(yintercept=c(6,10) ), color="#DFDFDF", size=1,
linetype="solid")
#step4 添加数据
g=g+geom_point(mapping= aes(time, value, fill=variable),
shape=21, color="NA", size=3,
data=df)+
scale_fill_manual('XX Gene', values = c('red', 'blue', 'purple'))+
geom_rect(data = NULL, # 中心圆环
aes(ymin = 5, ymax = 5.9),
xmin = -Inf, xmax = Inf,
fill = "white", colour = "white", size = 1,
linetype="solid")
return(g)
};draw(df)+coord_polar()
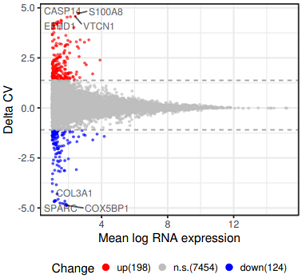
head(df3) #数据 7776 5
# mean_normal mean_sync mean delta status
#SPARC 0.02001691 3.917605 1.866243 -4.884748 down
#COX5BP1 0.01877168 4.004604 1.906797 -4.871463 down
#求平均数和标准差
m1=mean(df3$delta)
sd1=sd(df3$delta)
# 统计status列每个值的个数
tb=table(df3$status);tb
#down ns up
# 124 7454 198
#ggplot2 画图
g=ggplot(df3, aes( x= mean , y=delta, color=factor(status,levels=c('up','ns','down')) ) ) +
geom_point(alpha=0.5,size=0.1)+
labs( title="xxx", x='Mean log RNA expression', y="Delta CV" ) + theme_bw()+
theme(legend.box = "horizontal", # 图例,水平,标到底部
legend.key.size=unit(10,"pt"), #图例方块的高度
legend.position="bottom") +
scale_color_manual('Change', labels=c( paste0("up(",tb[3],")"),paste0('n.s.(',tb[2],')'),paste0("down(",tb[1],")") ), #图例文字
values=c("red", "grey",'blue') )+ #自定义颜色
#scale_shape(guide = guide_legend(title.position = "top", size=12)) +
geom_hline(aes(yintercept=m1+sd1*2), color="#aaaaaa", linetype="dashed")+
geom_hline(aes(yintercept=m1-sd1*2), color="#aaaaaa", linetype="dashed");g
# 为点添加文字注释
library(ggrepel)
dd_text=df3[which(df3$delta >m1+sd1*7 | df3$delta < (m1-sd1*8) ),];dim(dd_text)
g2=g+geom_text_repel(data=dd_text, aes(x=mean, y=delta, label=rownames(dd_text)),
color="black",size=3,alpha=0.6)+
guides(colour = guide_legend(override.aes = list(alpha = 1,size=2)))#放大图例的点
# 保存到文件
library(Cairo)
CairoPDF(file="03_RNA_Sync_vs_normal_HeLa_CV_changed_genes_v3.pdf",width=4,height=4)
print(g2)
dev.off()
可以使用 text()
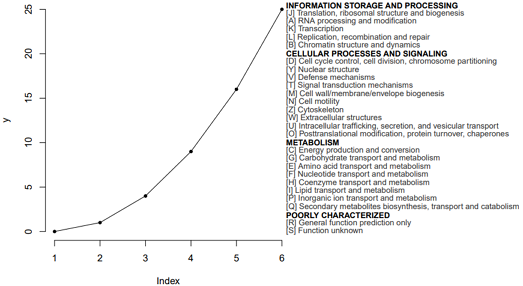
# 读取文件
text1=read.delim("fun.txt",header=FALSE) #该文件在文末
text1
len=nrow(text1); #29
b=20;
# 绘图
#tiff(filename="haha.tif",width=25,height=12,units="cm",compression="lzw",bg="white") #?
#png(file="aaaa.png",width=800,height=600);
#svg('aaaa.svg', width=8, height=6)
pdf('aaaa.pdf', width=10, height=6)
# 左边的图
par(fig=c(0,0.55,0,1),bty="n");
plot(c(0,1,4,9,16,25), type='o',ylab="y", pch=20)
# 右边的图: 文字
par(fig=c(0.4,1,0,1),bty="n", new=TRUE); #fig=c(x1, x2, y1, y2), 添加到新图 new = TRUE
plot(1:b,1:b,type="n", #type="n"不生成任何点和线,创建坐标系
xaxt="n",yaxt="n",
xlab="",ylab="");
sum=b+b/(2*len); #20.3448
for(i in 1:(len)){
if (i %in% c(1,7,18,27) ){
text(1,sum,text1[i,],adj=0,cex=0.8,font=2); #大号字体
}else{
text(1,sum,text1[i,],adj=0,cex=0.8, col='grey20')
}
sum=sum-b/(len); #一行的高度差不多是 20/1.6896
print(sum)
}
dev.off()
####### 补充材料
$ cat fun.txt
INFORMATION STORAGE AND PROCESSING
[J] Translation, ribosomal structure and biogenesis
[A] RNA processing and modification
[K] Transcription
[L] Replication, recombination and repair
[B] Chromatin structure and dynamics
CELLULAR PROCESSES AND SIGNALING
[D] Cell cycle control, cell division, chromosome partitioning
[Y] Nuclear structure
[V] Defense mechanisms
[T] Signal transduction mechanisms
[M] Cell wall/membrane/envelope biogenesis
[N] Cell motility
[Z] Cytoskeleton
[W] Extracellular structures
[U] Intracellular trafficking, secretion, and vesicular transport
[O] Posttranslational modification, protein turnover, chaperones
METABOLISM
[C] Energy production and conversion
[G] Carbohydrate transport and metabolism
[E] Amino acid transport and metabolism
[F] Nucleotide transport and metabolism
[H] Coenzyme transport and metabolism
[I] Lipid transport and metabolism
[P] Inorganic ion transport and metabolism
[Q] Secondary metabolites biosynthesis, transport and catabolism
POORLY CHARACTERIZED
[R] General function prediction only
[S] Function unknown
怎么让GO分析结果更高大上呢?
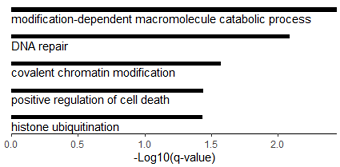
#数据源是 metascape 做的GO分析结果: http://www.metascape.org/
# install.packages('xlsx')
loadGOfromXLS=function(fname){
library(xlsx)
dat = read.xlsx(fname, sheetName = "Enrichment", encoding = 'UTF-8')
#dim(dat) #164 9
# (1)filter by p value
dt2=dat[which(dat$Log.q.value. < log10(0.05)),]
head(dt2[,c(1,2,3,6)])
#dim(dt2) #17 9
#colnames(dt2)
#1."GroupID" "Category" "Term" "Description" "LogP" "Log.q.value."
#7 "InTerm_InList" "Genes" "Symbols"
# (2)filter out duplicate terms
dt3=dt2[!duplicated(dt2[,'Description']),]
# (3) keep Summary only
dt4=dt3[grep('Summary',dt3$GroupID),]
return(dt4)
}
library(ggplot2)
GO_barplot=function(dt0){
dt0=dt0[order(-dt0$Log.q.value.),]
ggplot(dt0, aes(x=Description, y=-Log.q.value.
#, fill=Category
))+
geom_bar(stat="identity", width=0.15, fill="black")+
coord_flip()+
scale_x_discrete(limits=dt0$Description,labels = NULL )+
theme_classic()+
theme(legend.position="none")+
scale_y_continuous(expand = c(0,0))+ #去掉坐标轴两端的空白
annotate("text", x=seq(1, nrow(dt0))-0.3, y=0, #位置
hjust = 0, #文字左对齐
label= dt0$Description)+
labs(x="", y="-Log10(q-value)")+
theme( axis.ticks.y = element_blank(),
axis.line.y = element_blank(),
axis.text.y = element_blank() )
}
# read and plot
fname="F:\\c1_Figure\\Fig2\\vstVar_changed_HeLa\\down-313\\all.tfo2sgtfh\\metascape_result.xlsx"
dt4=loadGOfromXLS(fname)
GO_barplot(dt4)
# or plot using part of these items.
g2=GO_barplot(dt2[c(seq(1,8), 11,15,16,18,19,20),])
g2
## save to pdf
library(Cairo)
CairoPDF(file='F:/c1_Figure/GO_replot/06_CCNB1-pearson_cor-89genes.pdf',width=7,height=5)
g2
dev.off()
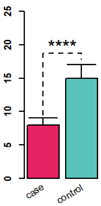
左: barplot 添加error bar;
右: line plot 添加error bar; 对坐标轴axis和轴标题、刻度标签间的距离、刻度长度的控制。
# R base 版 barplot
data = data.frame(mean = c(8, 15), sd = c(9, 17))
rownames(data) = c("case", "control")
par(lwd = 2)
xPos = barplot(data$mean, col =c('#e82364', '#5bc4ba'), #c("red", "blue"),
ylim = c(0, 25), axes = F, font = 2)
b=xPos
text(x=xPos, y=-1.2, labels=rownames(data),
xpd=TRUE, #允许绘制在绘图区外
adj=1, #adj=1右上对齐
srt=30) #倾斜30度
arrows(xPos[1], data$mean[1], b[1], data$sd[1], angle = 90) # error bar
arrows(xPos[2], data$mean[2], b[2], data$sd[2], angle = 90) # error bar
# 顶上的虚线
lines( x = c(b[1], b[1], b[2], b[2]), y = c( data$sd[1] * 1.05 , data$sd[2] * 1.1, data$sd[2] * 1.1, data$sd[2] * 1.05), lty = 2)
# 星号
text( x = b[1] + (b[2] - b[1]) / 2, y = data$sd[2] * 1.1, label = "****", cex = 2, adj = c(0.5, 0))
# 加坐标轴
axis(side = 2, lwd = 2, font = 2, cex = 1.5)
# R base 版: line plot
# 输入数据是均值和标准差
> head(dfB2)
mean sd
1 0.000000000 0.000000000
2 0.008797837 0.001955405
3 0.020597459 0.005301535
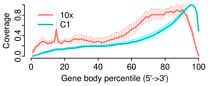
########## 3rd Edition: use mean and sd plot
library(Cairo)
CairoPDF("10x_c1.geneBodyCoverage.curves-3.pdf", width=5, height=3)
x=1:100
plot(NULL, NULL, type='l', xlim=c(0,100), ylim=c(0,1), axes = F,
mgp=c(1.5,0.5,0),
xlab="Gene body percentile (5'->3')", ylab="Coverage",lwd=0.8,col=icolor[1])
# 10x
icolor = colorRampPalette(c("#F8766D"))(10)
#
arrows(x0=seq(1,100), y0=dfA2$mean-dfA2$sd, x1=seq(1,100), y1=dfA2$mean+dfA2$sd,
angle=90, length = 0.015, col='#FEB1AB', lwd=0.5)
lines(seq(1,100),dfA2$mean, col=icolor[1], lwd=2)
# C1
icolor = colorRampPalette(c("#00BFC4"))(10)
#
arrows(x0=seq(1,100), y0=dfB2$mean-dfB2$sd, x1=seq(1,100), y1=dfB2$mean+dfB2$sd,
angle=90, length = 0.015, col='#02F8FE', lwd=0.5)
lines(seq(1,100),dfB2$mean, col=icolor[1], lwd=2)
#
legend(0,0.99, col=c('#F8766D', '#00BFC4'),lty=1, legend = c('10x', 'C1'), bty='n', lwd=2)
axis(side = 1, #x轴
lwd = 1, font = 1, cex = 1.5,
mgp=c(1.5,0.3,0), #标签文字与坐标轴的距离
tck=-0.04) #刻度线长度
axis(side = 2, lwd = 1, font = 1, cex = 1.5,mgp=c(1.5,0.3,0),tck=-0.04)
dev.off()
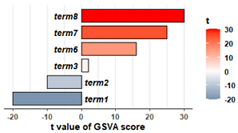
特点: 左右双向条形图。
# 模拟输入数据 GSVA的输出结果,包含name和t两列
rs1=data.frame(
name=c('term1', 'term2', 'term3', 'term4', 'term5', 'term6', 'term7', 'term8'),
t=c(-20,-10,2,5,15, 16,25,30)
)
head(rs1)
# 绘图函数
drawGSVA=function(data, n=10){
#n=3 #20
data_0 = rbind(
data[order(data$t,decreasing = T),][1:n,], #选择t值最极端的2*n个
data[order(data$t,decreasing = F),][1:n,])
# t的最值,用于优化坐标轴,默认注释掉,不用
#a = round(min(data_0$t)); b = round(max(data_0$t))
library(ggplot2)
# 画barplot
p=ggplot(data_0,aes(x=reorder(name,t),y=t,fill=t))+
geom_bar(stat = "identity",colour="black",width = 0.78,position = position_dodge(0.7))+
xlab("")+ylab("t value of GSVA score") +
# scale_y_continuous(breaks = seq(a,b,(b-a)/6))+
coord_flip() # 翻转xy轴
# 主题修饰
p=p+theme_bw()+
theme(panel.grid.major.y = element_blank(),panel.grid.minor.y = element_blank(),
axis.text.y = element_blank(),panel.border = element_blank(),
axis.ticks.y = element_blank(),axis.line.x = element_line(),
axis.text.x = element_text(face="bold"),axis.title.x = element_text(face="bold"))
# 加上文字
p2=p+geom_text(aes(y=ifelse(t>0,-1,1),label=name), #文字的y坐标。旋转后
hjust=ifelse(data_0$t>0,1,0), #按照t值正负,水平调整文字左右对齐
fontface=4,size=3.8
)+
scale_fill_gradient2(low="#366488",high="red",mid="white",midpoint = 0,name="t")+
# ylim(-60,60) + # 设置y轴的范围
theme(legend.text = element_text(face="bold"),legend.title = element_text(face="bold"),
# legend.position = c(0.9,0.3),
legend.direction = "vertical")
return(p2)
}
# 测试
drawGSVA(rs1, n=3)
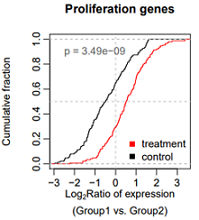
目标图片: Nat Commun. 2020 Widespread transcript shortening through alternative polyadenylation in secretory cell differentiation
Fig. 4. Single-cell analysis reveals short 3ʹUTRs in SCTs.
(c) Cumulative distribution function (CDF) curves comparing 3ʹUTR APA REDs in SCTs vs. VCTs (from Vento-Tormo et al. data) for genes showing 3ʹUTR shortening in the hESC model (blue curve, 685 genes from Fig. 2c) and all 3ʹUTR APA genes (black curve, 2379 genes). P-value (K–S test) for significance of difference between the two gene sets is indicated.
该图用来比较2个分布的异质性,曲线上升最快的区域对应的x轴区域,是数据集中区域。KS test用于检验2个分布的差异是否显著。
##########
# 累计密度曲线,并做K-S检验
##########
set.seed(2)
control=rnorm(150,-0.5,1)
treatment=rnorm(150, 0.5, 1)
#
hist(control, n=100)
hist(treatment, n=100)
#
pos1=min(c(control, treatment))
pos2=max(c(control, treatment))
## K-S test
rs.test=ks.test(control, treatment)
p=rs.test$p.value;p
p0=formatC(p, format = "e", digits = 2) #保留2位有效数字
p0
# plot
pdf("xx005.pdf", width=3.5, height=4)
plot(ecdf(control), col="black",
verticals = TRUE, do.points = FALSE, #画竖线,不画点
cex=0.4, xlim=c(pos1, pos2),
main="Proliferation genes",
xlab=expression( atop( paste("Log"["2"], "Ratio of expression"),
"(Group1 vs. Group2)") ),
mgp=c(3,0.6,0),
ylab="Cumulative fraction")
abline(v=0, col="grey", lty=2)
abline(h=0.5, col="grey", lty=2)
lines(ecdf(treatment), col="red",
verticals = TRUE, do.points = FALSE,
cex=0.4)
#添加图例
legend('bottomright',
box.col = NA, bg = NA,
legend=c("treatment", "control"), # text in the legend
col=c("red","black"), # point colors
pch=15) # specify the point type to be a square
#添加p值
text(-2.5,0.9, adj=0, labels = paste("p", "=", p0), col="#666666")
dev.off()
# Fail: 想把P设置斜体,同时显示P=1.2x10-9的形式,显示x而不是e,使用上标
#text(-2.5,0.9, adj=0, labels = expression(paste(italic("P"), " =", p0)))
#text(-1.6,0.9,adj=0, labels=p0)
# end
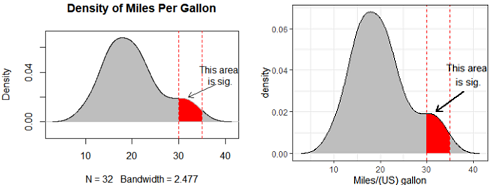
####### density plot with annotation
#########################
# Part I: R base 参考
#########################
set.seed(1)
draws <- rnorm(100)^2
dens <- density(draws)
plot(dens)
q2 <- 2
q65 <- 6.5
qn08 <- -0.8
qn02 <- -0.2
x1 <- min(which(dens$x >= q2))
x2 <- max(which(dens$x < q65))
x3 <- min(which(dens$x >= qn08))
x4 <- max(which(dens$x < qn02))
with(dens, polygon(x=c(x[c(x1,x1:x2,x2)]), y= c(0, y[x1:x2], 0), col="red"))
with(dens, polygon(x=c(x[c(x3,x3:x4,x4)]), y= c(0, y[x3:x4], 0), col="grey"))
# R base 模仿
d <- density(mtcars$mpg, n=2024) #n越大,后续定位越精细
plot(d, main="Density of Miles Per Gallon",
type="n",ylim=c(-0.01,0.07))
# graphical parameters such as xpd, lend, ljoin and lmitre can be given as arguments.
polygon(d, col="grey", border=NA) # Filled Density Plot
lines(d, col="black") #make outer line clear
# 涂灰色:范围
i1 <- min(which(d$x >= 30));
i2 <- max(which(d$x <= 35))
polygon(x=c( d$x[c(i1, i1:i2, i2)] ), y= c(0, d$y[i1:i2], 0), col="red",
lwd=1, border = "grey")
abline(v=c(30,35), lty=2, col="red")
# 箭头和注释
arrows(x0 = 38, y0 = 0.03, x1 = 32, y1 = 0.02,
# x0, y0,x1,y1 支持一次设置多个值,同时画多个箭头
length=0.1, # length 默认值为0.25, 为了调整不同箭头的大小
code=2, #code : 调整箭头的类型,一共有1,2(默认),3, 共3种类型
#通用的参数,col , lty ,lwd 等
lwd=1, lty=1,
angle=30) #箭头张开角度
text(39,0.038, labels="This area \nis sig.")
#########################
# Part II
#########################
# ggplot2 参考 https://www.jianshu.com/p/c8a886ba5803
#http://www.sthda.com/english/wiki/ggplot2-area-plot-quick-start-guide-r-software-and-data-visualization
# ggplot 模仿
library(ggplot2)
df=data.frame(x=mtcars$mpg)
head(df)
dat <- with(density(df$x), data.frame(x, y))
head(dat)
#
dat1<-dat[dat$x<(30),] #left
dat2<-dat[dat$x>35,] #right
ggplot()+
geom_density(data=df,aes(x=x),fill="red")+
geom_area(data=dat1,aes(x=x,y=y),fill="grey")+
geom_area(data=dat2,aes(x=x,y=y),fill="grey")+
geom_line(data=dat, mapping=aes(x,y), size=0.1, color="black")+ #smooth outer line
geom_vline(xintercept = c(30,35), linetype=2, color="red")+ # vertical line
geom_segment(data = NULL,
aes(x=38,y=0.03, xend=32, yend=0.02), size = 0.8,
arrow = arrow(length = unit(0.3, "cm")))+ #箭头
annotate("text", x =39, y = 0.038, label = "This area \nis sig.") + #文字注释
labs(x="Miles/(US) gallon")+
theme_bw()
第一版没有error bar,模仿 fig.3c
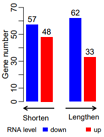
ct=c(33, 62, 48, 57)
#
dt2=data.frame(
length=c("lengthen","lengthen", '', "shorten","shorten"),
exp=c('up','down', '', 'up','down'),
#value=c(54,84,NA,85,83)
value=c(ct[1:2],NA, ct[3:4])
)
dt2
pdf("xx_barPlot.pdf", width=2.8, height=3.8)
par(mgp=c(1.4,0.5,0) )
posX=barplot( rev(dt2$value), col=rev(c("red",'blue','white',"red",'blue')),
ylim=c(0,70), border=NA, ylab="Gene number" )
text(x=posX, y=rev(dt2$value)+4, labels=rev(dt2$value) )
#text(x=posX-0.3, y=-6, offset = 1,
# srt = 30, xpd = TRUE, cex=0.9,
# labels=rev(dt2$exp) )
# arrows
arrows(3.5,-5,6,-5, length = 0.08,
col='black', xpd = T, code=2, lwd=2)
text(x=5, y=-12, xpd = TRUE, cex=0.9,
labels="Lengthen" )
#
arrows(0,-5,2.5,-5, length = 0.08,
col='black', xpd = T, code=1, lwd=2)
text(x=1, y=-12, xpd = TRUE, cex=0.9,
labels="Shorten" )
#
legend(-2.5,-15,xpd = TRUE, horiz=T,
#box.lwd=3,
text.width=2.5,
x.intersp=0.5,
border=NA, cex=0.8,
#title="RNA", title.adj=1,
fill=c('white','blue',"red"), legend = c('RNA level','down', 'up'), bty='n' )
dev.off()
注: 今天发现pdf硬盘占用比png小很多,尝试在网页嵌入pdf图片。缺点是有黑边,不知道怎么去掉。
难点: 对x坐标重排序,对y坐标对数尺度; 组内连线;
library(ggplot2)
library(dplyr)
# 生成数据 ID, treatment, time和values
set.seed(20211103)
mydata = data.frame(
ID = c(rep(1:10, 2), rep(11:20, 2)),
treatment = rep(c("v", "p"), each = 20),
time = rep(c("Before", "After", "Before1", "After1"), each = 10),
values = c(rnorm(10, mean = 100, sd = 60),
rnorm(10, mean = 155, sd = 60),
rnorm(10, mean = 105, sd = 60),
rnorm(10, mean = 100, sd = 60))
)
mydata$values=10^( mydata$values / ( max(mydata$values)-min(mydata$values) ) *4 )
mydata
summary(mydata$values)
# ID代表受试者的编号;
# treatment有两个水平,v代表疫苗组,p为安慰剂组;
# time分为before和after,分别指代治疗前和治疗后;
# values代表血液中的抗体水平。
str(mydata)
#'data.frame': 40 obs. of 4 variables:
# $ ID : int 1 2 3 4 5 6 7 8 9 10 ...
#$ treatment: chr "v" "v" "v" "v" ...
#$ time : chr "Before" "Before" "Before" "Before" ...
#$ values : num 27.781 21.426 1.857 0.712 9.931 ...
#png("00test.png", width=72*3*0.8, height=72*4*0.8, res=72)
#svg("00test.svg", width=3*0.8, height=4*0.8)
pdf("00test.pdf", width=3*0.8, height=4*0.8, useDingbats=F)
ggplot(mydata, aes(time, log10(values), fill=time))+
#scale_y_log10()+ #纵坐标取对数
geom_point(shape=21, size=3, show.legend = F)+
scale_x_discrete(limits = c("Before", "After", "Before1", "After1"),
labels = c("Before", "After", "Before", "After")) + # 修改x轴刻度上的标签
scale_y_continuous( breaks = seq(-1,6,1),
labels = 10^seq(-1,6,1) ) + # 修改x轴刻度上的标签
scale_fill_manual(values = c("indianred3", "steelblue", "indianred3", "steelblue")) + # 修改颜色
geom_hline(yintercept = 2, linetype = "dashed") + # 添加水平虚线
geom_line(data = filter(mydata, treatment == "v"), aes(group = ID)) + # 添加疫苗组的点对点线条
geom_line(data = filter(mydata, treatment == "p"), aes(group = ID)) + # 添加安慰剂组的点对点线条
xlab( "mRNA-xxx Placebo") + # 修改x轴的标签
ylab("Anti-RBD Antibody (U/ml)") + # 修改y轴的标签
theme_classic()
dev.off()
来源: 群友提问。paper: //todo
后续整理版: 微信公众号 雷达图
喜欢这个GO富集结果可视化效果,距离圆心越远表示越显著,颜色越深表示基因个数越多。
模拟数据文件:
$ cat dustbin/8dat.csv
Desc,id,log10padj,Count
IL-17,1,9.87,11
Rheumatoi,2,7.27,9
Viroal,3,7.16,9
TNF signal,4,5.54,8
Cytokine,5,5.25,11
Chemokine,6,4.98,9
Glycolysis,7,4.69,6
Legionell,8,3.786,5
Lipid and,9,3.782,8
Amoebiasi,10,3.782,6
画图:
# 尝试 v3: 给一个函数版本 //todo
# 两个主要变量:
# 几边形? 10
# 分成几层? 4
# 最大半径? 10
# 多边形数据: 获取圆上的n个点的坐标
# rotate: 为了美观,整体逆时针旋转的弧度
getPointsOnCircle=function( radius=1, npoints=10, center=c(0,0), rotate=pi/2){
angle=seq(0, 2*pi, length.out=npoints+1)
xx=center[1] + radius*cos(angle+rotate)
yy=center[2] + radius*sin(angle+rotate)
return(data.frame(x=xx, y=yy))
}
# 画多边形
drawPolygon = function(radius, npoints=10, linetype=1, size=0.5){
geom_polygon(data= getPointsOnCircle(radius, npoints), aes(x, y),
linetype=linetype,
size=size, fill="#FFFFFF00", color="#AAAAAA")
}
# (原点到各个顶点)放射线 顶点坐标
getRadialLines = function(maxR=10, npoints=10){
arr=pi/2 + seq(0, 2*pi, length.out=npoints+1);
x=c()
y=c()
for(i in arr){
x=c(x, c(0, maxR)*cos(i))
y=c(y, c(0, maxR)*sin(i))
}
return(data.frame(x, y))
}
# 读取数据
dat=read.csv("dustbin/8dat.csv"); head(dat)
dat
# 10个方向上的富集分析: 10边形
dat2=dat
dat2$angle=pi/2+seq(0, 2*pi, length.out=11)[1:10]
dat2$x=dat2$log10padj * cos(dat2$angle)
dat2$y=dat2$log10padj * sin(dat2$angle)
dat2
library(ggplot2)
g1=ggplot()+ xlim(-12, 12) + ylim(-12, 12)+
drawPolygon(10)+drawPolygon(7.5)+drawPolygon(5)+drawPolygon(2.5)+ #雷达图 几层多边形
geom_path(data=getRadialLines(10, 10), mapping=aes(x,y), color="#AAAAAA", linetype=1)+ #雷达图放射直线
geom_polygon(data=dat2, aes(x, y), fill="#CB5656",alpha=0.5, color="#CF0015",size=1 )+ #填充不规则多边形
#
geom_point(data=dat2, aes(x,y, fill=Count), size=5, shape=21, color="black", stroke=0.5)+ #添加大点
scale_fill_gradient(low="white", high="#00B858", breaks=seq(5, 11, 2) )+ #点的渐变色。控制图例数字 breaks+labels
#
geom_text(data=data.frame(x=-0.8, y=seq(0,10,2.5) ), aes(x, y, label=y),
color='black', angle=0, hjust=1, vjust=1 )+ # 仿y坐标值刻度 tick
annotate(geom="text",x=0.2, y=5,
label="-log[10]*italic(P)*adj", parse=T,
color="black", angle=90, vjust=1 )+ #仿y轴名字 label
#
geom_text(data=dat2, aes(x=11*cos(angle), y=11*sin(angle), label=Desc), color="black")+# 每条边的文字
#
coord_equal()+ #保证x和y轴等长
theme_void(base_size = 12)+ #去掉坐标轴、边框
theme(
legend.position = "top",
legend.justification = "left", #靠左对齐
legend.key = element_rect(fill = "black") #不起作用 //todo
)
g1
pdf("dustbin/radar.pdf", width=4, height=4)
g1
dev.off()
# 输出pdf矢量图,然后可以使用 Illustrator 编辑文字:添加换行、调整文字位置、字号等。
欢迎互相切磋,共同进步: 秋秋号 314649593, 请备注大名、来意。
秋秋群: 生物信息与R语言 187923577 禁止营销活动,否则飞机票。
bioToolKit is part of 生物慕课网 www.biomooc.com